Research in the Beehive
The BEEHIVE develops tailored models and methods using approaches from statistics and machine learning in order to study, deconstruct, visualize, predict, modify, and engineer biomedical systems.

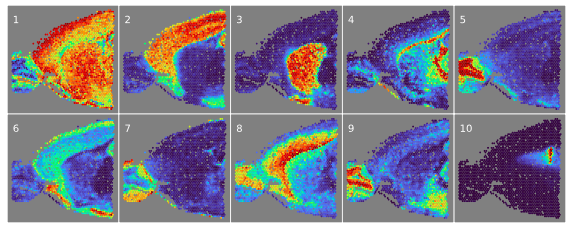
Spatial single cell analyses
Measurements of biological systems have both noise and systematic bias, and often the analytical goal is to identify biologically-meaningful low-dimensional substructure within a high-dimensional space with structured samples, such as time-series data, single-cell RNA-sequencing, spatial transcriptomics samples, or CRISPR screens. We build model-based approaches to gain access to interesting biological phenomena that would be otherwise missed.

Electronic health records
Models. We have built a number of models based on Gaussian processes for electronic health records. First, we built a multi-output Gaussian process model to capture 24 different patient covariates across time using a linear model of coregionalization (LMC) to capture the relationship between each covariate [Cheng et al. 2020]. We adapted this model to handle medical interventions, such as medication, using latent force models that for a limited time modify the values of specific covariates, such as the application of beta blockers that modify systolic and diastolic blood pressure for 4-6 hours [Cheng et al. 2020b]. We extended these ideas to a hierarchical Gaussian process to allow different categorical variables such as race or sex across patients [Cui et al. 2022].
Decision making. We applied off-policy reinforcement learning to a number of clinician-in-the-loop decision-making tasks. We first used reinforcement learning to find an optimal policy to remove patients from mechanical ventilation [Prasad et al. 2017]. Next, we applied reinforcement learning (RL) to schedule lab tests for patients, saving a large number of blood tests and timing the tests to correspond with diagnostic actionability [Cheng et al. 2019]. We also applied RL to the challenge of electrolyte repletion in hospital patients, improving dosage and methods to maintain appropriate levels of electrolytes [Prasad et al. 2022].
Methods. We have developed a number of methods for use in EHR data. With Finale Doshi-Velez, we developed a method to identify policies that could be evaluated given the existing held-out patient data [Prasad et al. 2020]. We built a nested RL method that assumes that there are two sets of patients, with slightly different dynamics, that share a reward function, such as electrolyte repletion in patients that also include patients with end-stage renal disease and cannot process electrolyte supplements as well as patients with healthy kidneys [Mandyam et al. 2022]. We built a Bayesian inverse reinforcement learning method to estimate the posterior distribution of the objective function of the task [Mandyam et al. 2023].

Live-cell imaging
Canine kidney cells with a wound inflicted on the right hand side [Toettcher Lab]
Blue: Low ERK signaling; Green: High ERK signaling; Red: background