Methods for cellular behavior analysis in live-cell imaging
Our preprint on cellular behavior analysis in TCR T cells & cancer cell live-cell imaging co-cultures is out! This three-year collaboration led by postdoc Archit Verma is in collaboration with Alex Marson & Julia Carnevale, with cellular segmentation & tracking by David Van Valen & his amazing team! [bioRxiv].
Incucyte live cell imaging is ubiquitous, but from this complex data cancer immunologists typically plot one thing: the number of red pixels in the well, which is a proxy for the cancer cell coverage (RFP marks cancer cell nuclei). From [Carnevale et al. 2022]:
A lot of interesting signal is left on the cutting room floor. For example, proliferation (rates) in cancer cells, cancer cell death, which is quite rare in much of these data, and amazing pack hunting behaviors of the modified T cells (I’m struggling with the videos — I’ll get help and get back to you!).
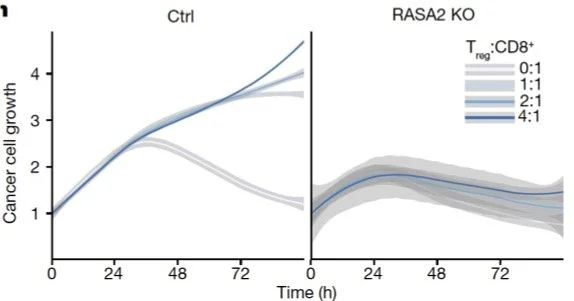
We teamed up with Alex Marson and Julia Carnevale’s Labs, who imaged TCR T cells co-cultured with RFP+ A375 tumor cells under three conditions: Safe harbor knockout (SH KO; control), RASA2 KO [Carnevale et al., 2022], CUL5 KO [Liao et al., 2024]. Each well was imaged every four minutes at 10X magnification on Sartorius Incucyte for 72 hrs. Images include brightfield, RFP (marking cancer cell nuclei) channels.
Then, the Van Valen Lab developed Caliban to segment and track each cell (green are T cells, red are cancer cells):
With the masked, tracked cells, we went to work to develop Occident. We were curious how well the RFP markers captured cancer cell number; we found that RFP lags as a proxy for cancer cell numbers.
We found that T cell proliferation increased in the two beneficial KO T cells, in the CUL5 KO T cells in particular.
We can study differences in cancer cell division events (lower in beneficial KO T cells) and average T cell speed (faster in beneficial KO T cells).
We found that the number of T cells attached to cancer cells reduces the likelihood that the cancer cell will proliferate, with the beneficial KO T cells having greater effects on proliferation reduction.
Cancer cell and T cell morphology changes dramatically depending on state. These changes are visible in the brightfield imaging – active interacting T cells are larger and change to less circular shapes. Cancer cell begin to aggregate together when interacting with T cells.
While the number of T cell--cancer cell interactions increased similarly across time, these interactions & their effects were modulated by the CRISPR KOs. For example, the amount of time a T cell remained attached to a cancer cell (as estimated by a negative binomial and Markov model separately) was highest in RASA2 KO T cells.
Even more exciting, the speed of cancer cells decreased after interactions with T cells, as did their overall size (indicating stress).
Most thrilling is that we can identify active T cells based on relative cell size and morphology, and watch T cells activate (differentially based on condition) after interacting with cancer cells.
With a Markov model, we deconvolved the situation when, in frame at time t-1, there is one cancer cell and one T cell in a small window, and in frame at time t there is one cancer cell and two T cells. We were able to quantify how often this doubling of T cells attacking a cancer cell was due to proliferation or due to recruitment.
In summary, we found that, compared to the SH KO control condition, TCR T cells with the RASA2 KO have a longer dwell time and cripple cancer cells more effectively this way, whereas TCR T cells with the CUL5 KO proliferated more frequently upon activation, adding more T cells to the fight.
With five new collaborations in the works, and a paper characterizing the differences in condition using explainable AI already accepted as an oral presentation at #PSB2025 (lead by high school senior Marcus Blennemann), look for future work in this space, and try out Caliban and Occident on your own Incucyte data! More phenotypes and analyses added regularly.
Feedback welcome! And please play with these data! There is a lot more signal there.
Thank you to Bioimage Archive for hosting these Incucyte image data -- this is a new thing for them, and they have been so kind in working through the details of submission (link coming soon!)! 🎉